Evidence for Chlamydia in Crohns Disease
Herbert J Van Kruiningen, Aaron W Hayes, Antonio Garmendia, Junru Cui, Francine B de Abreu, Gregory J Tsongalis and Jean Frederic Colombel
DOI10.4172/2472-1891.100015
Herbert J Van Kruiningen1*, Aaron W Hayes1, Antonio Garmendia1, Junru Cui1, Francine B de Abreu2, Gregory J Tsongalis2 and Jean Frederic Colombel3
1Department of Pathobiology and Veterinary Science, University of Connecticut, Storrs, CT, USA
2Department of Pathology, Dartmouth-Hitchcock Medical Centre and Geisel School of Medicine at Dartmouth, Lebanon
3Division of Gastroenterology, Icahn School of Medicine at Mount Sinai, New York, NY, USA
- *Corresponding Author:
- Herbert J Van Kruiningen
Department of Pathobiology and Veterinary Science
University of Connecticut, Storrs, CT, USA
Tel: (860) 486-0837
E-mail: herbert.vankruiningen@uconn.edu
Received date: October 23, 2015; Accepted date: January 22 2016; Published date: February 02, 2016
Citation: Herbert J Van Kruiningen, Aaron W Hayes, Antonio Garmendia, et al. Evidence for Chlamydia in Crohn’s Disease. Int J Dig Dis. 2016, 2:1. doi:10.4172/2472-1891.100015
Abstract
Chlamydia causes enteritis in young pigs and calves, and granulomas within intestinal lymphatics. Recently we called attention to the lymphangitis and lymphatic obstruction that occurs in Crohn’s disease. We searched resected tissues from patients for evidence of chlamydia and sera for evidence of previous exposure to chlamydia. Immunohistochemistry and real-time PCR were employed to seek chlamydia in preserved tissues. In the IHC, antibody to C. trachomatis served as primary antibody; in the PCR, primers specific for Chlamydiaceae were employed. Commercial ELISA kits measured anti-chlamydia IgG and IgA against C. trachomatis antigen in sera derived from a population of patients different from that which yielded the tissue specimens. IHC revealed focal positive staining for chlamydia in tissues of 5 of 19 patients. Positive reacting cells occurred within dense inflammation, in sparsely scattered macrophages in the submucosa and subserosa. Tissues from 3 of 22 control subjects were positive. Real-time PCR done on ileal, colonic, and regional lymph node tissues revealed evidence of chlamydia in 3 of 33 patients. Serology for anti-chlamydia IgG revealed 2 positive values in 24 patients, while serology for anti-chlamydia IgA revealed 4 positives among the 24 patients, and 1 positive in the 15 controls. One patient and one control had both elevated IgG and IgA titers. The 4 patients with elevated IgA titers were from a single family of 6 with Crohn’s disease, which had been previously described. Additional consideration needs to be given to the chlamydia species, including those of animal origin, which leave behind little evidence of their previous involvement.
Keywords
Crohn’s disease; Inflammatory bowel disease; Chlamydia
Introduction
Recently we studied the lymphatics in resection specimens from patients with Crohn’s disease [1,2]. Employing serial sections of granuloma-obstructed lymphatics or of lymphatics that were distended with lymphocytes, we showed physical continuity between the former and the latter [2]. Given the importance of this lymphangitis, we began to search for viral or bacterial antigens that might be responsible [3].
In 2013, we took new interest in a previously-described chlamydial enteritis of young pigs, one that shows its greatest effect in the distal ileum [4]. After oral inoculation, Chlamydia suis invade and damage villous epithelial cells, then induce inflammation in villous lamina propria, and finally enter and damage lymphatic endothelium, the latter accompanied by intra- and extralymphatic inflammation [5,6]. One consequence of this localized intestinal infection is the production of granulomas that obstruct mucosal, submucosal, muscularis, and serosal lymphatics [4,6]. The finding that C. suis replicates what occurs in Crohn’s disease raises the question: Is there evidence that human patients with Crohn’s disease harbor or have been exposed to C. suis or other Chlamydia?
C. suis is closely related to Chlamydia trachomatis [7,8]. The thought that the latter might be responsible for Crohn’s disease had been examined earlier. Serologic studies over the years yielded equivocal results, as many studies showing antibody as those failing to find any (Table 1).
| Author | Year | Country | Ab | Antigen | Test | N + | Total | % | Ctr+ | Ctr Total | Ctr % + |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Rodaniche et al[34] | 1943 | US | LGV | SN | 0 | 4 | 0 | ||||
| Swarbricket al [35] | 1979 | England | IgG | C trachomatis | MicroFA | 4 | 54 | 7.4 | 7 | 50 | 14 |
| Taylor-Robbinsonet al [36] | 1979 | England | C trachomatis | Micro FA | 8 | 55 | 14.5 | 5 | 23 | 22 | |
| Schulleret al [37] | 1979 | Holland | IgG | C trachomatis | Micro FA | 38 | 55 | 69 | 1 | 50 | 2 |
| Mardhet al [38] | 1980 | Sweden | IgG | C trachomatis | IIF | 83 | 107 | 78 | 38 | 50 | 76 |
| Gump et al [39] | 1981 | England | LGV(L2) | IIF | 36 | 60 | 60 | 34 | 60 | 57 | |
| Elliot et al [9] | 1981 | England | C trachomatis | Micro FA | 0 | 62 | 0 | 3 | 160 | 2 | |
| Ordaet al [10] | 1990 | Israel | IgG | C trachomatis | IIP | 14 | 15 | 93 | 4 | 15 | 26 |
| Ordaet al [10] | 1990 | Israel | IgA | C trachomatis | IIP | 5 | 15 | 33 | 1 | 15 | 6 |
| McGarityet al [16] | 1991 | England | IgG | C trachomatis | ELISA | 7 | 48 | 15 | 14 | 48 | 29 |
Table 1: Serologic studies seeking chlamydia antibody in patients with Crohn’s disease.
Elliot et al seemed to have closed the door on this issue when they could not culture chlamydia from the tissues of patients (N=14), could not demonstrate chlamydia in tissue sections by FA (N=5), and could show antibody responses in only 3 of 62 patients [9]. Orda on the other hand found IgG responses to C. trachomatis in 14 of 15 patients (93%) [10].
There having been no search specific for C. suis and some variability in the methods and results of antibody testing of patients with Crohn’s disease, we examined tissues from patients by immunohistochemistry and real-time polymerase chain reaction (PCR), and sera from patients by ELISA.
Materials and Method
Formalin-fixed paraffin-embedded tissue blocks from surgically resected diseased ilea and proximal colons from 19 CD patients from France and the United States were examined. These were transverse full thickness cuts, 1–3 per subject. Lennard-Jones criteria [11]. and histopathology were used for establishing diagnosis. Cases for study were selected for transmural obstructed lymphatics, and granulomas. Patients were 7 men and 12 women; aged 18–46 years (mean age 26). Formalin-fixed tissues from control patients were examined as well, 15 men and 7 women (mean age 48 years). Control tissues were from patients that had undergone colectomy for adenocarcinoma, ulcerative colitis, indeterminate colitis, diverticulitis, mesenteric venous thrombosis or gunshot trauma. Duration of disease ranged from 1 month to 9 years from the time of diagnosis. This study was approved by the Institutional Review Board of the University of Connecticut.
Primary antibodies for immunohistochemistry were obtained from commercial sources; mouse monoclonal anti-Chlamydia trachomatis LPS antibody [EVH-1] was employed to label chlamydia (this antibody cross-reacts with LPS of other chlamydia), (Abcam, Cambridge, MA). Microsections of tissues 4-μm thick were placed on positively charged glass slides and deparaffinized in xylene (Allegiance Healthcare, McGaw Park, IL, USA), rehydrated in a graded series of ethanol and rinsed with distilled water. Immunohistochemistry was performed as previously described [12]. Antigen retrieval was performed by using proteinase K for 10 minutes at room temperature. This was followed by washing in phosphate-buffered saline (Dako, Carpinteria, CA); then endogenous peroxidase activity was blocked by incubation in 3% H2O2 for 10 min., followed by application of Animal Free Blocker (Vector, Burlingame, CA) for 13 minutes at room temperature. After another wash in Dako wash buffer, tissue sections were incubated with the primary antibody, at a dilution of 1:1500 for 60 min at room temperature. After a short wash in Dako wash buffer, tissue sections were incubated for 30 min with HRP-labeled polymer conjugated with secondary antibody (EnVisionTM+ Dual Link System-HRP, Dako). Following a final wash in Dako wash buffer, sections were developed with Nova Red (Vector) for 2 min. After washing in warm tap water, the slides were counterstained with hematoxylin, then dehydrated, cleared and coverslipped. Appropriate negative and isotype controls (omitting primary antibody) were run with each batch of 20–40 test sections. Positive controls were C. suis- infected pig intestine.
Twenty micron thick sections from the paraffin-embedded tissues of 33 CD patients (the 19 from the IHC study and an additional 14 from France and the US) and the same 22 controls were submitted for real-time PCR. Genomic DNA was extracted using the Gentra PureGene Kit for tissues (Qiagen, Valencia, CA) and quantified with the NanoDrop 1000 spectrophotometer (Thermo Fisher Scientifc Inc., Waltham, MA, USA). Samples were screened for Chlamydia spp. on the AB 7500 Fast Real Time PCR System (Applied Biosystems, Foster City, CA, USA) using the 23S rRNA gene based Chlamydiaceae family specific method, which includes primers Ch23S-F (5’-CTGAAACCAGTAGCTTATAAGCGGT-3’), Ch23S-R (5’-ACCTCGCCGTTTAACTTAACTCC-3’) and probe Ch23S-P (FAM-CTCATCATGCAAAAGGCACGCCG) [13]. Each run included 2 positive controls, 2 negative controls, and samples. Positive controls used were sections of C. suis-infected pig intestine. Negative controls were urine samples previously screened for Chlamydia trachomatis (CT), Trichomonas vaginalis (TV) and Neisseria gonorrhoeae (NG) using a test developed and validated at the Translational Research Laboratory at the Dartmouth- Hitchcock Medical Center (Lebanon, NH, USA) [14]. Negative samples were negative for CT and TV, and positive for NG. Both samples and controls were run in duplicate. PCR was performed using 2x primer/probe mix, 2x TaqMan GTXpress Master Mix (Applied Biosystems) and 12ng of DNA. Cycling conditions were 50°C for 2 minutes, 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. The positive samples showed a cycle threshold of <38.00. Samples positive for Chlamydia spp. were confirmed by melt curve analysis using the SmartCycler II (Cepheid, Sunnyvale, CA). PCR was performed using 25x primers, 2x SsoFastTM EvaGreen® Supermix (Bio-Rad, Hercules, CA) and 12ng of DNA. Cycling conditions were 95°C for 5 minutes, followed by 40 cycles of 95°C for 5 sec and 60°C for 30 sec. The melt curve was determined at 65-95°C, 0.2°C per 1 sec. Positive samples showed a specific melting temperature (Tm) of approximately 84°C.
Sera from a separate set of CD patients (N=24) and sex- and age-matched controls (N=15) from northern France were tested for anti-chlamydia antibodies. ELISA was performed using Anti- Chlamydia trachomatis IgG Human ELISA Kit, and Anti-Chlamydia trachomatis IgA Human ELISA Kit (Abcam, Cambridge, MA) according to the manufacturer’s instructions. Briefly, all materials were equilibrated to room temperature prior to use. For initial detection of specific antibodies serum samples were diluted 1:100 with sample diluent as indicated in manufacturer’s instructions. Limited cross-reactivity data are available for the kit used. Positive samples were titrated further. Diluted samples were added into wells in duplicates and incubated at 37°C for 1 hour. The wells were washed five times with 1X washing solution. After the last wash Chlamydia trachomatis anti-IgG HRP or anti-IgA HRP conjugate was added and incubated at room temperature for 30 minutes. The plates were washed as above and TMB substrate solution was added into each well and incubated at room temperature in the dark for 15 minutes, when a stop solution was added to the wells. The absorbance was measured at 450nm in a Biotek Synergy HT microplate reader (Biotek Winosski, VT). The kit manufacture’s criteria were used to validate the test and score the samples as positive or negative.
Results
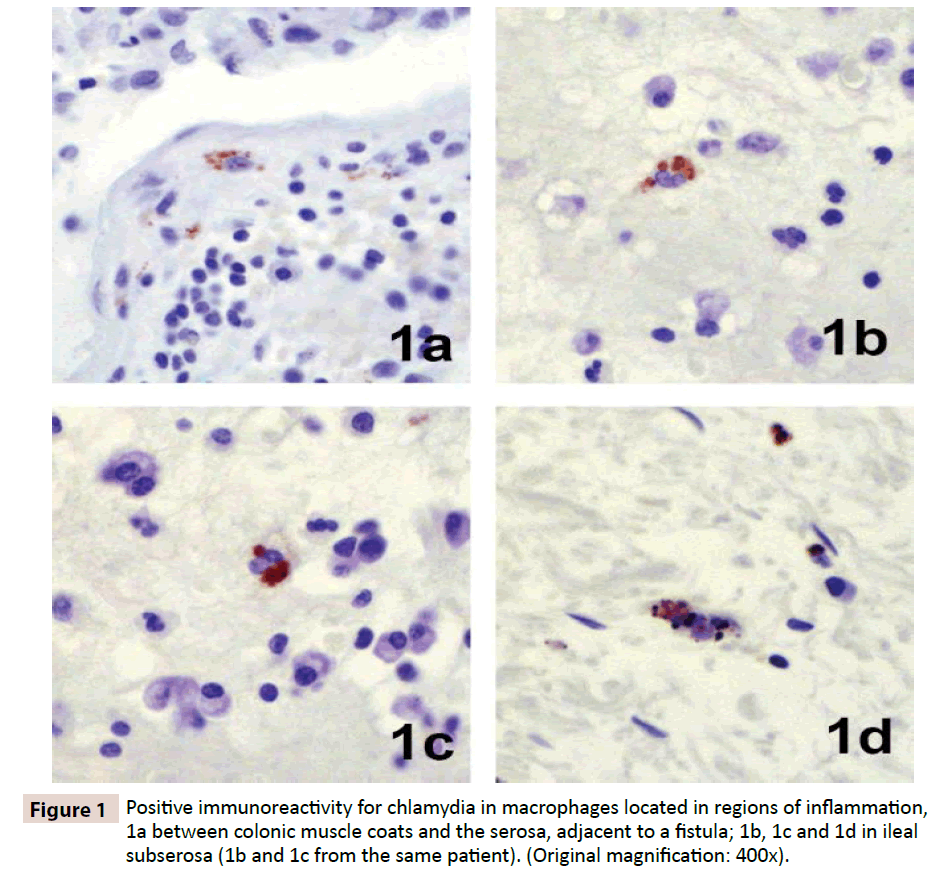
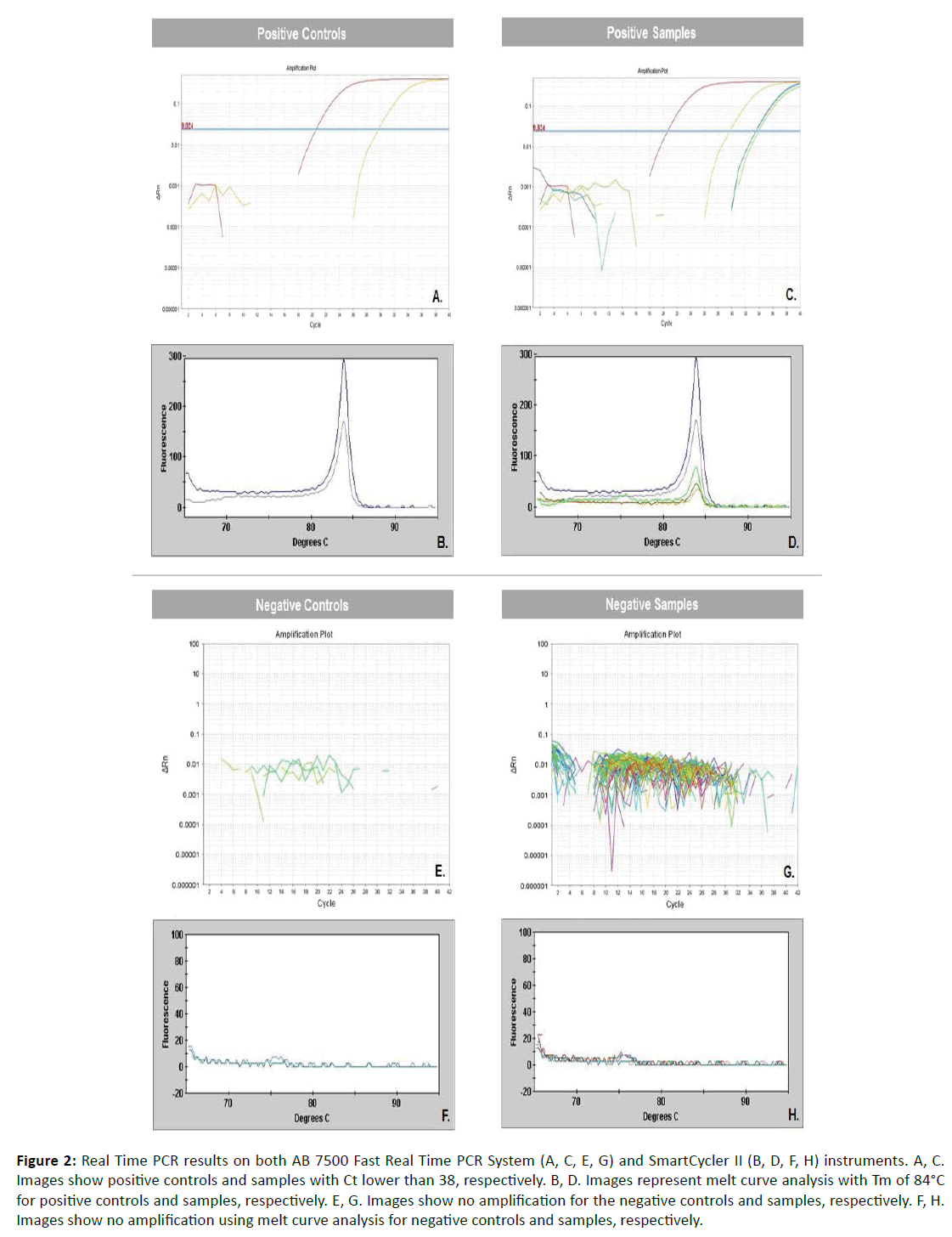
Immunohistochemistry revealed focal positive staining for chlamydia in the tissues of five of 19 patients with Crohn’s disease. Positive reacting cells occurred within dense inflammation, in sparsely scattered macrophages in the submucosa and sub-serosa (Figure 1). Focal positive staining also occurred in 3 of 22 controls, one resection for ulcerative colitis, one for indeterminate colitis, and one for mesenteric thrombosis. Real-time PCR performed on DNA extracted from ileal, colonic and regional lymph node tissues revealed evidence of Chlamydia spp. in three of 33 patients tested, two ileums (Figure 2) and one lymph node, and in none of 22 controls.
Figure 2: Real Time PCR results on both AB 7500 Fast Real Time PCR System (A, C, E, G) and SmartCycler II (B, D, F, H) instruments. A, C. Images show positive controls and samples with Ct lower than 38, respectively. B, D. Images represent melt curve analysis with Tm of 84°C for positive controls and samples, respectively. E, G. Images show no amplification for the negative controls and samples, respectively. F, H. Images show no amplification using melt curve analysis for negative controls and samples, respectively.
ELISA serology for anti-chlamydia IgG revealed two positive values in sera of 24 CD patients tested versus one positive in 15 controls. ELISA serology for anti-chlamydia IgA revealed four positives in the 24 patients tested and one positive in the 15 controls. One patient, number 9, and one control, number 19, had both elevated IgG and IgA titers. Importantly, the four patients who had elevated chlamydia-specific IgA antibodies were from a family of 6, previously described, all of whom had CD [15]. Duplication of this testing, done with a second ELISA kit carrying a different lot of antigen, confirmed these findings. Further titration of the positive sera detected IgA antibody titers of 1/400 in one patient serum and 1/200 in another patient. A similar titer of 1/200 was detected in one control serum.
Discussion
There is no strong association here between the presence of Chlamydia spp. and Crohn’s disease. McGarity et al, using a chlamydia plasma probe and PCR, found no evidence of C. trachomatis in 5 resection specimens and 5 biopsies from CD patients [16]. Employing a PCR-EIA method for the 16S rRNA gene of C. trachomatis on mucosal biopsies Chen et al found one positive colon in 11CD patients and one positive colon in 37 controls [17].
Why then should we care about five positives here, three positives there, and four positives in another population? These positive data are small in number; however, among the microbial agents known to induce a lymphangitis, there are few that localize to the intestine and induce granulomatous obstruction of lymphatics [5, 6].
One needs to recognize that by the time our specimens were acquired from patients, the disease had been present for many years. Experimental studies in which young pigs, calves, and nonhuman primates were inoculated with chlamydia have shown that the organisms cannot be demonstrated in the tissues after 12 weeks [5,18]. The chlamydia are known to persist in tissues in a form that cannot be readily demonstrated [19-21]. These dormant forms can be reactivated. When C. trachomatis strains were inoculated into rectal mucosa in monkeys, the LGV strain generated lymphoid follicles and giant cells within six weeks [18]. Intracytoplasmic inclusions were seldom seen after three weeks. Inflammatory follicles that were created (tertiary lymphoid organs) were well-developed with germinal centers, and persisted [18], however immunohistochemistry for chlamydia was negative after 10-12 weeks.
The duration of antibodies to chlamydia is variable, and waning with time [18,22,23]. And, they only indicate exposure during some point in the lifetime of the subject. Pathogen-specific serum IgA has been recognized as an important indicator of a number of viral infections [24] (Epstein-Barr virus [25,26], rubella [27], varicella-zoster [28], cytomegalovirus [29]). In chlamydial infections, elevated levels of serum IgA have been regarded as an indication of either deep-seated infection [30] or active chlamydial disease [24,31].
That all four of our IgA positives were from one family with CD is remarkable. Did the family have point source contact with C. trachomatis of other organ systems? Were they exposed to a chlamydia other than C. trachomatis that cross-reacted serologically? Or does this finding reveal important information about the etiology of Crohn’s disease? In 1990, and again in 1992, the medical records of each member of this family were extensively reviewed, and each of the living were interviewed in the presence of the family gastroenterologist [15]. At that time, the time the sera were acquired, there was no evidence of other chlamydia- associated disease, leaving us to conclude that the elevated serum IgA titers to chlamydia reported here are a reflection of intestinal disease. We do wish to acknowledge that in our testing there may have been cross-reactivity with chlamydia other than C. trachomatis, perhaps chlamydia of animal origin. The real-time PCR was broadly specific for Chlamydiaceae 23SrRNA.
In the search for an etiologic agent in Crohn’s disease, the chlamydia are sound candidates; some species induce lymphoid follicles, some induce granulomas, and some do both. Most importantly, however, some penetrate the intestinal mucosa and damage lymphatics, as in the pig disease [4-6] and in calves [22,32,33]. Additional consideration needs to be given to the Chlamydia spp. – however, defining the initiator of Crohn’s disease may be impossible, given the unknown time from (a presumed) gastroenteritis in childhood or young adulthood to clinical manifestation of the effects of damaged ileal or ileocolonic lymphatics.
Acknowledgement
The authors acknowledge the support of Laurent Dubuquoy, INSERM Lille, France and Dr. AB West, Yale University School of Medicine New Haven, CT for obtaining surgical specimens, Denise Long at the University of Connecticut for the immunohistochemistry, and Jeffrey Magin for technical support. This research was funded by the Association F Aupetit and the University of Connecticut Research Foundation.
IRB Approval
This study was approved by the Institutional Review Board of the University of Connecticut.
References
- Sura R,Colombel JF, Van Kruiningen HJ (2011)Lymphatics, tertiary lymphoid organs and the granulomas of Crohn's disease: An immunohistochemical study. Aliment PharmacolTher 33:930-939.
- Van Kruiningen HJ, Hayes AW, Colombel JF (2014) Granulomas obstruct lymphatics in all layers of the intestine in Crohn's disease. APMIS: actapathologica, microbiologica, etimmunologicaScandinavica 122:1125-1129.
- Magin WS, Van Kruiningen HJ, Colombel JF (2012)Immunohistochemical search for viral and bacterial antigens in Crohn's disease. Journal of Crohn's & colitis.
- Rogers DG, Andersen AA (1996) Intestinal lesions caused by two swine chlamydial isolates in gnotobiotic pigs. Journal of veterinary diagnostic investigation: official publication of the American Association of Veterinary Laboratory Diagnosticians, Inc 8:433-440.
- Rogers D, Andersen A (2000) Intestinal lesions caused by a strain of chlamydia suis in weanling pigs infected at 21 days of age. J Vet Diagn Invest 12:233-239.
- Guscetti F, Schiller I, Sydler T, Heinen E, Pospischil A (2009) Experimental enteric infection of gnotobiotic piglets with chlamydia suis strain s45. Veterinary Microbiology 135:157-168.
- Donati M, Di Francesco A, Delucca F, Di Paolo M, Battilani M, et al. (2011) Antibody-neutralizing activity against all urogenital chlamydia trachomatis serovars in chlamydia suis-infected pigs. FEMS Immunol Med Microbiol 61:125-128.
- Everett KD (2000) Chlamydia and chlamydiales: More than meets the eye. Veterinary Microbiology 75:109-126.
- Elliott PR, Forsey T, Darougar S, Treharne JD, Lennard-Jones JE (1981)Chlamydiae and inflammatory bowel disease. Gut 22:25-27.
- Orda R, Samra Z, Levy Y, Shperber Y, Scapa E (1990) Chlamydia trachomatis and inflammatory bowel disease--a coincidence? J R Soc Med 83:15-17.
- Lennard-Jones JE (1989) Classification of inflammatory bowel disease. Scand J Gastroenterol24 170:2-6.
- Cartun R, VanKruiningen H, Pedersen C (1993)Animmunohistochemical search for infectious agents in Crohn's disease. Mod Pathol 6:212-219.
- Borel N, Regenscheit N, Di Francesco A, Donati M, Markov J, et al. (2012) Selection for tetracycline-resistant chlamydia suis in treated pigs. Veterinary Microbiology 156:143-146.
- AbouTayoun AN, Burchard PR, Caliendo AM, Scherer A, Tsongalis GJ (2015) A multiplex pcr assay for the simultaneous detection of chlamydia trachomatis, neisseriagonorrhoeae, and trichomonasvaginalis. ExpMolPathol 98:214-218.
- Van Kruiningen HJ, Colombel JF, Cartun RW, Whitlock RH, Koopmans M, et al. (1993) An in-depth study of Crohn's disease in two frenchfamilies. Gastroenterology 104:351-360.
- McGarity BH, Robertson DA, Clarke IN, Wright R (1991) Deoxyribonucleic acid amplification and hybridisation in Crohn's disease using a chlamydial plasmid probe. Gut 32:1011-1015.
- Chen W, Li D, Wilson I, Chadwick VS (2002) Detection of chlamydia pneumoniae by polymerase chain reaction-enzyme immunoassay in intestinal mucosal biopsies from patients with inflammatory bowel disease and controls. Journal of gastroenterology and hepatology 17:987-993.
- Quinn TC, Taylor HR, Schachter J (1986) Experimental proctitis due to rectal infection with chlamydia trachomatis in nonhuman primates. The Journal of Infectious Diseases 154:833-841.
- Cheema MA, Schumacher HR, Hudson AP (1991)Rna-directed molecular hybridization screening: Evidence for inapparent chlamydial infection. Am J Med Sci 302:261-268.
- Hudson AP, McEntee CM, Reacher M, Whittum-Hudson JA, Taylor HR (1992)Inapparent ocular infection by chlamydia trachomatis in experimental and human trachoma. Curr Eye Res 11:279-283.
- Hogan RJ, Mathews SA, Mukhopadhyay S, Summersgill JT, Timms P (2004) Chlamydial persistence: Beyond the biphasic paradigm. Infection and Immunity 72:1843-1855.
- Ronsholt L (1978) Chlamydia psittaci infection in danish cattle. ActapathologicaetmicrobiologicaScandinavica Section B, Microbiology 86B:291-297.
- Quinn TC, Goodell SE, Mkrtichian E, Michael D. Schuffler, San-Pin Wang, et al. (1981) Chlamydia trachomatis proctitis. The New England Journal of Medicine 305:195-200.
- Cevenini R, Sarov I, Rumpianesi F, Donati M, Melega C,et al. (1984) Serum specific iga antibody to chlamydia trachomatis in patients with chlamydial infections detected by elisa and an immunofluorescence test. Journal of clinical pathology 37:686-691.
- Evans AS, Niederman JC (1982)Ebv-iga and new heterophile antibody tests in diagnosis of infectious mononucleosis. American journal of clinical pathology 77:555-560.
- Ho HC, Ng MH, Kwan HC, Chau JC (1976) Epstein-barr-virus-specific iga and igg serum antibodies in nasopharyngeal carcinoma. Br J Cancer 34:655-660.
- Hornsleth A, Leerhoy J, Grauballe P, Spanggaard H (1975) Persistence of rubellavirus-specific immunoglobulin m and immunoglobulin a antibodies: Investigation of successive serum samples with lowered immunoglobulin g concentration. Infection and Immunity 11:804-808.
- Levy E, Sarov I (1981) Detection of specific iga antibodies in serum of patients with varicella and zoster infections. Intervirology 15:103-110.
- Sarov I, Levy E, Aymard M, Chardonnet Y, Bosshard S, et al. (1982) Detection of virus-specific iga antibodies in serum of kidney transplant patients with recurrent cytomegalovirus infection by enzymeimmuno and radioimmunoassay techniques. ClinExpImmunol 48:321-328.
- Scheel O, Anestad G (1989) Significance of immunoglobulin a titres in the diagnosis of urogenital chlamydial infections. Eur J ClinMicrobiol Infect Dis 8:726-728.
- Kumamoto Y, Sato T, Hiroi M, Hashizume S, Nakata H,et al. (1993) Assessment of chlamydia trachomatis-specific iga and igg serum antibodies in genitourinary chlamydia trachomatis infection--comparative study between hitazyme and ipazyme. KansenshogakuZasshi 67:315-330.
- Doughri AM, Altera KP, Storz J, Eugster AK (1973) Electron microscopic tracing of pathogenetic events in intestinal chlamydial infections of newborn calves. ExpMolPathol 18:10-17.
- Doughri AM, Yong S, Storz J (1974) Pathologic changes in intestinal chlamydial infection of newborn calves. American journal of veterinary research 35:939-944.
- Rodaniche E, Kirsner JB, Palmer W (1943)The relationship between lymphogranulomavenereum and regional enteritis: An etiologic study of 4 cases with negative results. Gastroenterology 1:687-689.
- Swarbrick ET, Kingham JG, Price HL,Blackshaw AJ, Griffiths PD et al. (1979) Chlamydia, cytomegalovirus, and yersinia in inflammatory bowel disease. Lancet 2:11-12.
- Taylor-Robinson D, O'Morain CA, Thomas BJ, Levi AJ (1979) Low frequency of chlamydial antibodies in patients with Crohn's disease and ulcerative colitis. Lancet 1:1162-1163.
- Schuller JL, Piket-van Ulsen J, Veeken IV, Michel MF, Stolz E (1979) Antibodies against chlamydia of lymphogranuloma-venereum type in Crohn's disease. Lancet 1:19-20.
- Mardh PA, Ursing B, Sandgren E (1980) Lack of evidence for an association between infection with chlamydia trachomatis and Crohn's disease, as indicated by micro-immunofluorescence antibody tests. ActapathologicaetmicrobiologicaScandinavica Section B, Microbiology 88:57-59.
- Gump D, Caul E, Eade O, Greenberg H, Kapikian A,et al. (1981) Lymphocytotoxic and microbial antibodies in Crohn's disease and matched controls. Antonie Van Leeuwenhoek 47:455-464.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences